Automated Machine Learning with Radiomics for Predicting Chronicity of Pulmonary Nodules in Patients with Nontuberculous Mycobacterial Lung Infection
Images




Abstract
Objective and Hypothesis: This study aimed to create a machine learning model to differentiate acute and chronic pulmonary nodules using radiomics-based analysis. Distinguishing between acute and chronic nodules on computed tomography (CT) is essential for patient management but remains challenging due to absence of standardized imaging criteria. We hypothesized that radiomic features could predict nodule acuity.
Materials Methods: We retrospectively analyzed 110 adult subjects with non-tuberculous-mycobacterial respiratory-infection with at least two chest CT scans between 2005 and 2021. Acute nodules were those initially present and subsequently resolved, while chronic nodules persisted beyond 30 days on follow-up scans. A total of 260 acute and 249 chronic nodules were individually segmented by a radiologist and radiology resident, with the overlapping segments extracted for radiomics analysis; 112 radiomic features were extracted. A test set of 108 nodules was assessed blinded by four radiologists; their performances were compared to the AI model. Recursive-feature-elimination and permutation-importance were used to reduce overfitting, resulting in eight final features used for model development. An auto-machine learning package developed the final predictive model. Test performance metrics between the model and the individual radiologists were compared using McNemar’s test and the area under the receiver operating curves (AUCs) for the model and the individual radiologists were compared using the DeLong test.
Results: The most accurate model was an ensemble model with sensitivity of 0.65, specificity of 0.92, positive predictive value of 0.88, negative predictive value of 0.75, and AUC of 0.88. The test performance metrics were significantly greater than two of the radiologists (P=0.011 and 0.020) and the AUC was significantly greater than all the radiologists (P value range:<0.0001–0.048).
Conclusions: This study demonstrates the feasibility of a machine learning model for predicting pulmonary nodule acuity using radiomics. The final model achieved an AUC of 0.88, significantly outperforming four radiologists.
Keywords: Nontuberculous mycobacterium, Machine learning, Radiomics, Pulmonary disease
Introduction
Nontuberculous mycobacterium (NTM) are pervasive organisms increasingly implicated in global respiratory morbidity and mortality. Evaluation of pulmonary changes related to NTM-induced lung disease (NTM-LD) via chest computed tomography (CT) scans is crucial for both diagnostic precision and therapeutic monitoring. In particular, the recognition of specific manifestations of NTM-LD is pivotal to clinical management. For example, patients with primarily nodular and bronchiectatic changes visible on CT imaging generally exhibit improved responses to antimycobacterial therapies, and patients with cavitary lesions or consolidations tend to respond more poorly to antimycobacterials.1 Further supporting this notion, patients achieving full resolution of lung nodules on high-resolution CT (HRCT) scans after a six-month treatment course are more likely to experience successful therapeutic outcomes, and patients with persistent nodules are more likely to experience treatment failure or relapse.2
While the tracking and evaluation of radiographic alterations are crucial for understanding NTM-induced lung disorders (NTM-LD), the nuanced assessment of evolving pulmonary nodule features can be laborious and subject to inter-observer variability. Various classification methods and radiologic features of disease for NTM-LD nodules have been suggested, particularly for distinguishing NTM-LD from Mycobacterium tuberculosis (TB) infection, although none have achieved universal acceptance.3,4
Artificial intelligence (AI) applications in medical imaging are increasingly becoming an integral part of clinical practice, revolutionizing the healthcare landscape in the process. These AI solutions often equal or outperform traditional computer-aided diagnostic methods. Specifically, deep convolutional neural networks (DCNNs) have demonstrated efficacy in classifying patients with active TB versus healthy individuals through chest X-rays and distinguishing between NTM-LD and TB-related pulmonary conditions using chest CT scans.5 Radiomics is another advanced imaging technique that can be particularly useful when dealing with smaller datasets and can describe and quantify human-recognizable features of imaging data, such as shape and pixel intensity features. Radiomics data may then be used for predictive or classification tasks.
Unlike DCNNs that often require large training datasets, radiomics can function efficiently on smaller data sets. The feature-based approach of radiomics also makes it more interpretable, allowing clinicians to trace back the outcomes to specific, understandable imaging features. This interpretability can be crucial for gaining clinical trust and for medical decision-making, especially in complex cases where the pathophysiological relevance of extracted features may be better understood. Therefore, radiomics offers an approach that may be more suited to certain diagnostic challenges.
This study hypothesizes that a radiomics-based model has the potential to reliably forecast the activity status of CT-identified NTM-LD nodules over a minimum timeframe of 30 days, with non-inferiority to the diagnostic accuracy of radiologists.
Materials and Methods
This retrospective study received approval from the Institutional Review Board (IRB), with a waiver for informed consent owing to its retrospective nature and minimal risk to patients.
Data Sets
Patients were recruited from the Johns Hopkins Center for Nontuberculous Mycobacteria and Bronchiectasis and were diagnosed with NTM-LD between 2005 and 2021, in accordance with standard clinical guidelines.
Chest CT scans available on our institutional picture archiving and communication system (PACS) were reviewed. Exclusion criteria consisted of patients with either one or no available CT scans, insufficient scan quality, or lack of suitable nodules for annotation. Both contrast-enhanced and non-contrast CT scans, including those from external institutions uploaded to our PACS, were considered.
Imaging Evaluation
A radiologist specializing in thoracic imaging with 9 years of experience, blinded to clinical information, assessed the axial CT images. Images were evaluated using lung window settings (window width: 1400 HU; window level: -500 HU) to identify and categorize non-fully-calcified pulmonary nodules. Acute and chronic nodules were characterized based on their stability or resolution in subsequent scans. Acute nodules were defined as those present on the initial scan but which were no longer apparent within 30 days of follow-up. Chronic nodules were defined as those present on the initial scan that persisted beyond 30 days.
We selected a subset of the data consisting of 57 acute pulmonary nodules and 51 chronic pulmonary nodules, which a group of four radiologists (Readers 1-4) labeled in a blinded fashion. These same nodules were used to create the testing data set (108 nodules) and the remaining nodules were used to create the training data set (401 nodules), with no patient overlap between the two.
Segmentation, Radiomics, and Automated Machine Learning
Manual segmentation of all identified nodules was performed by a radiologist with 2 years of post-graduate experience and a third-year radiology resident, using 3D Slicer — an open-source software. The Dice Similarity Coefficient (DSC) was used to quantify the spatial overlap between the individual segmentations, and the overlapping areas were used for radiomic feature extraction.
Radiomic features were extracted and normalized using Pyradiomics (version 3.1.0).6 One-hundred-twelve radiomic features (comprising all features available in the base Pyradiomics library) were extracted. Dimensionality reduction techniques were employed to select a subset of optimized features for modeling; specifically, recursive feature elimination was employed with a Random Forest algorithm to identify 30 key radiomic features, which generated an initial model. Feature importances from these 30 were subsequently analyzed, and features that detracted from model performance were excluded. The model was subsequently recreated on the features with optimal performance.
The selected radiomic features were then analyzed using the Autogluon auto-machine learning package (version 0.8.2) to develop predictive models.7 During initial training, Autogluon constructs an ensemble of various models optimized for the chosen evaluation metric. From this ensemble, we extracted the highest-performing model based on the area under the receiver operating curve (AUC) perfor- mance. The evaluation metric was set to the AUC. The hyperparameters were configured to include bagging with 10 folds, 6 bagging sets, and 3 stacking levels. Training was limited to 120 minutes. Training was conducted on a GPU for computational efficiency. Additional hyperparameters were automatically optimized by Autogluon. Autogluon natively selects validation datasets from the training set.
Statistical Analysis
Model performance was assessed by accuracy (AC), sensitivity (SE), specificity (SP), positive predictive value (PPV), negative predictive value (NPV) and AUC on the testing dataset. The differences between the model’s SE and SP were compared to the results from the testing data of each radiologist individually using McNemar’s test and the differences between the model’s and radiologists’ AUCs were compared using the DeLong test, both with an alpha of 0.05. The contribution of individual features to the model was first assessed via feature ranking and subcategorization from recursive feature elimination. Individual feature importance was assessed via permutation shuffling with 5 shuffle sets. Statistical significance of feature importance was assessed via a t-test with the hypothesis: importance = 0, vs the (one-sided) alternative: importance > 0 with an alpha of 0.01. Model confidence scores for individual label examples were derived from the predicted probabilities of the model.
Results
Nodule Segmentation and Model Configuration
A total of 509 pulmonary nodules—comprising 260 acute and 249 chronic nodules—were segmented. Interobserver agreement between the two radiologists responsible for the segmentation yielded an average DSC of 0.89, with a standard deviation of 0.07. The final machine learning architecture employed was a Neural Net Fast AI model; bagging techniques were utilized to augment the model’s generalization capabilities.
Diagnostic Performance
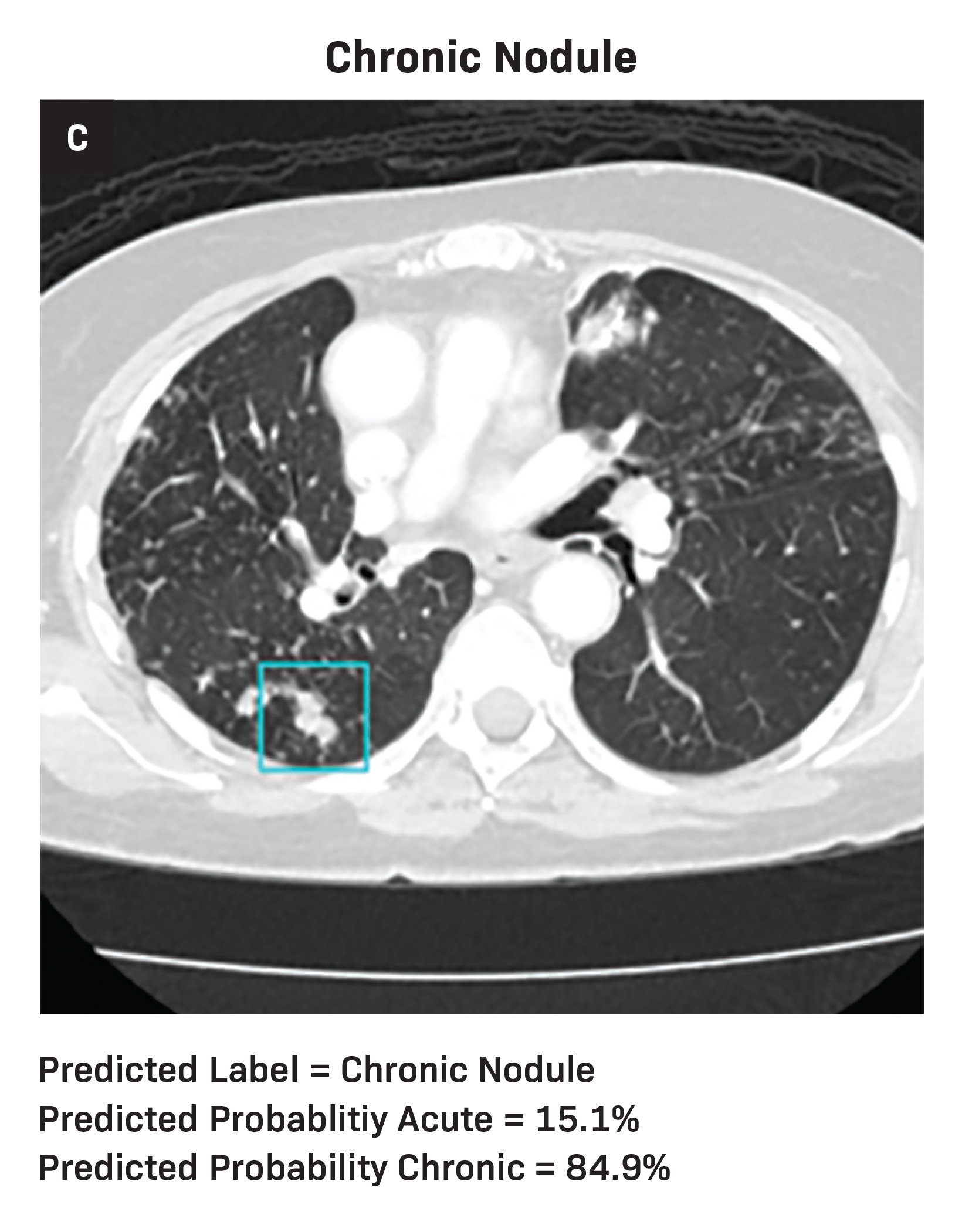
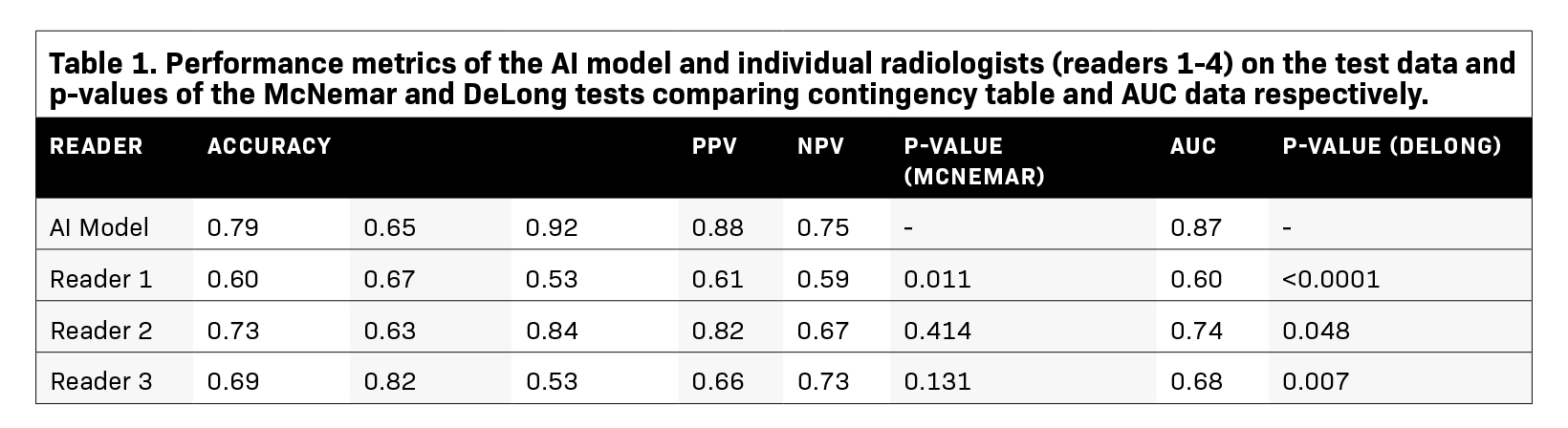
Diagnostic metrics for the AI model and individual radiologists are summarized in Table 1. Visual examples of both accurately and inaccurately labeled nodules, accompanied by model-derived confidence scores as predicted probabilities, are presented in Figure 1.
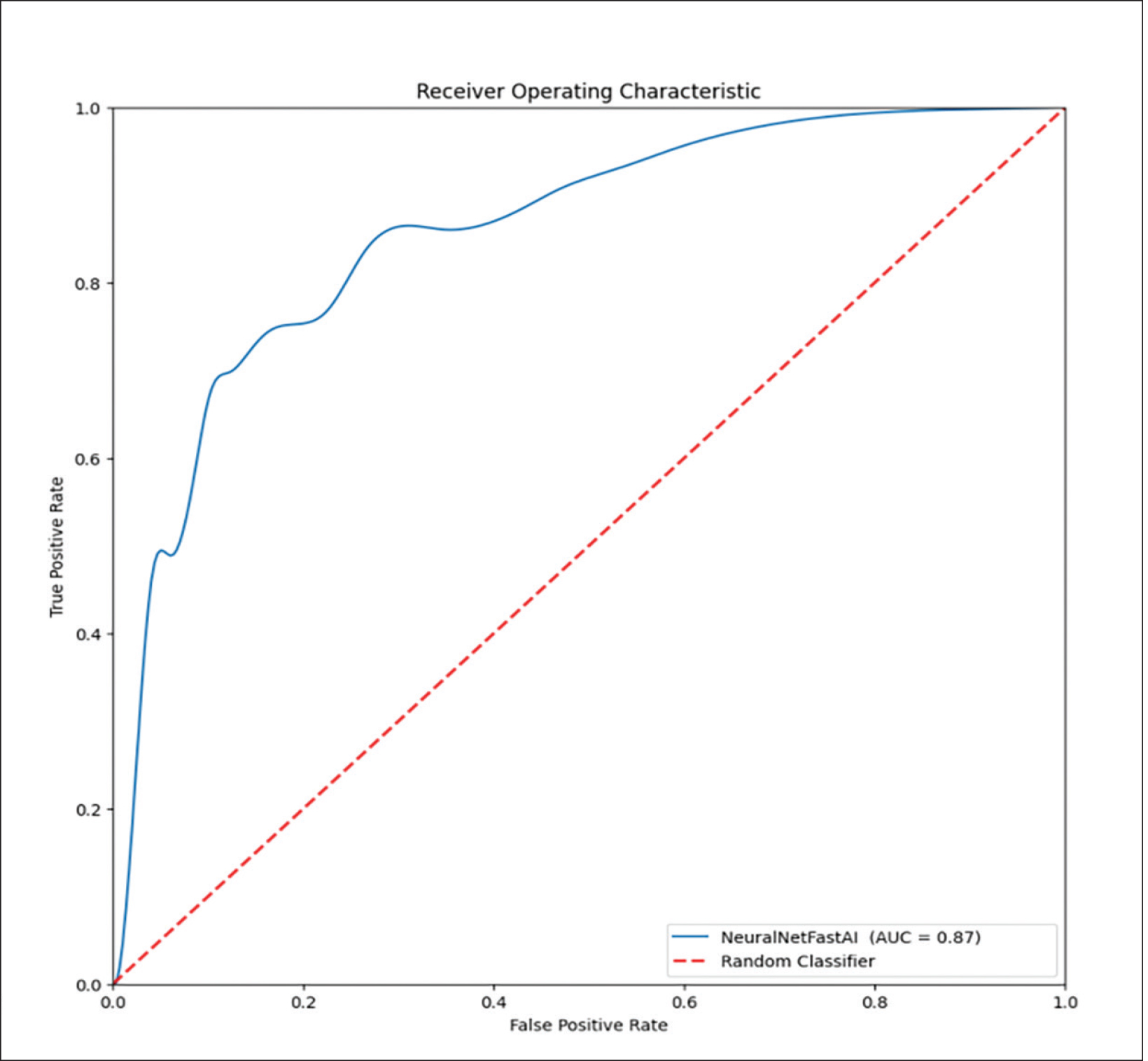
McNemar’s test identified significant differences in the classification performances between the AI model and two of the radiologists (Readers 1 and 4, P=0.011 and 0.020 respectively). Specifically, the SEs of the AI model and the two readers were similar (SeAI=0.65, SEReader 1=0.67, SEReader 4=0.61); however, the SP of the AI model was approximately 50% greater than the two radiologists (SPAI=0.92, SPReader 1=0.53, SPReader 4=0.67, Table 1). Additionally, the AI model’s AUC outperformed all of the radiologists (AUCAI=0.87, AUCReader 1=0.60, AUCReader 2=0.74, AUCReader 3=0.68, AUCReader 4=0.64, P-value range:<0.0001–0.048, Table 1, Figure 2). The model had overall high performance in the classification of chronic nodules with PPV of 0.88 and moderate performance in the classification of acute nodules with NPV of 0.75. The radiologists achieved a wide range of PPVs (0.61-0.82) and NPVs (0.61-0.73).
Feature Importance and Contributions
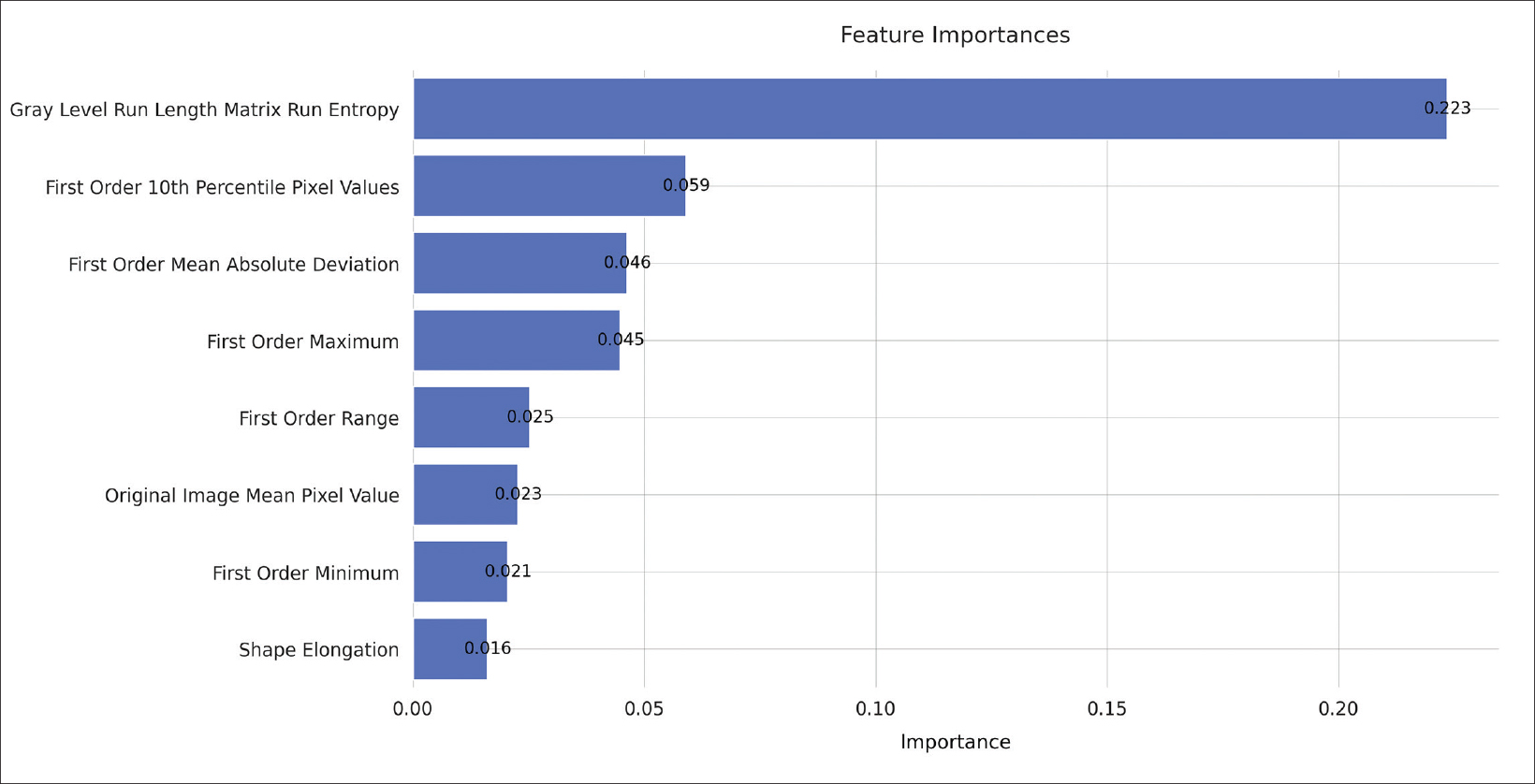
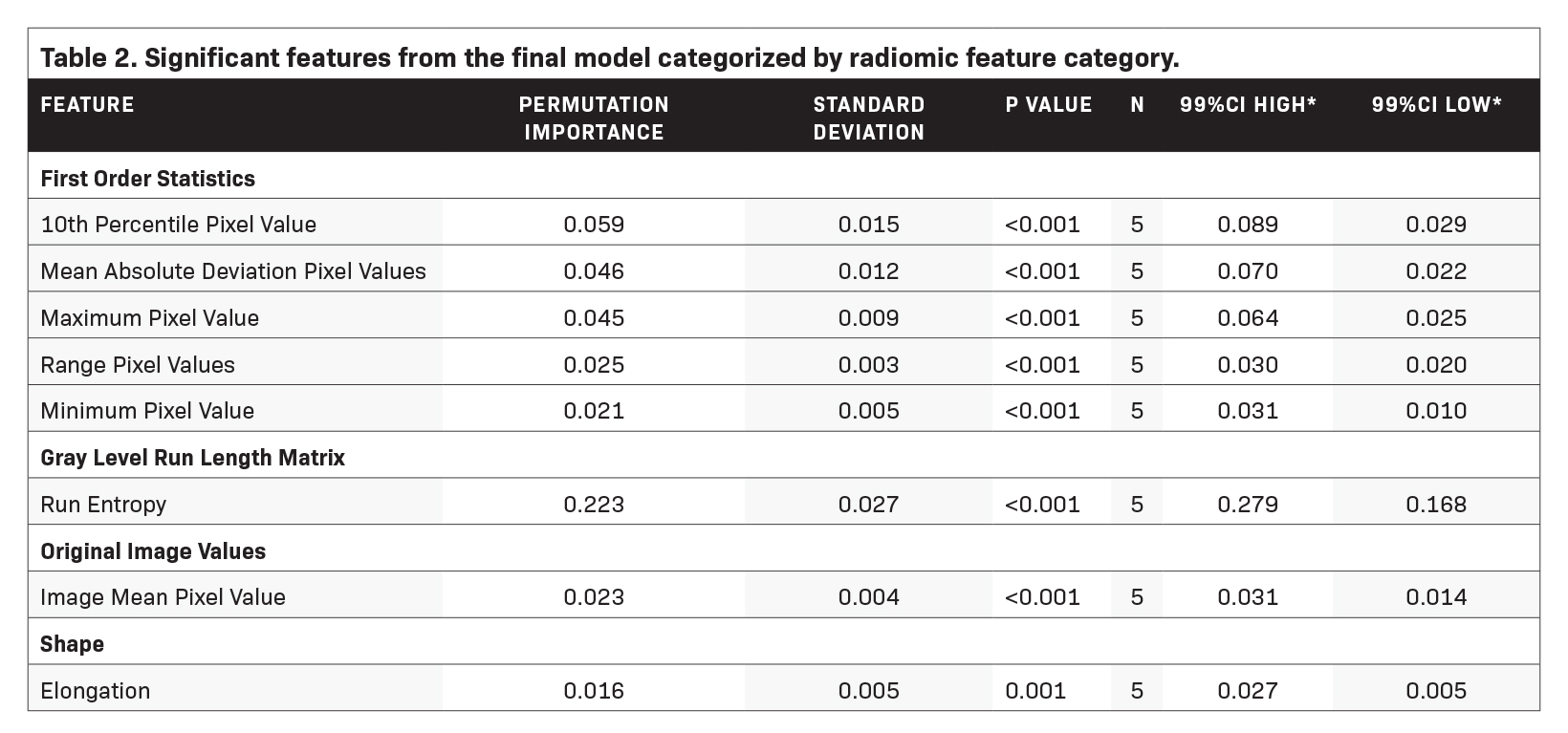
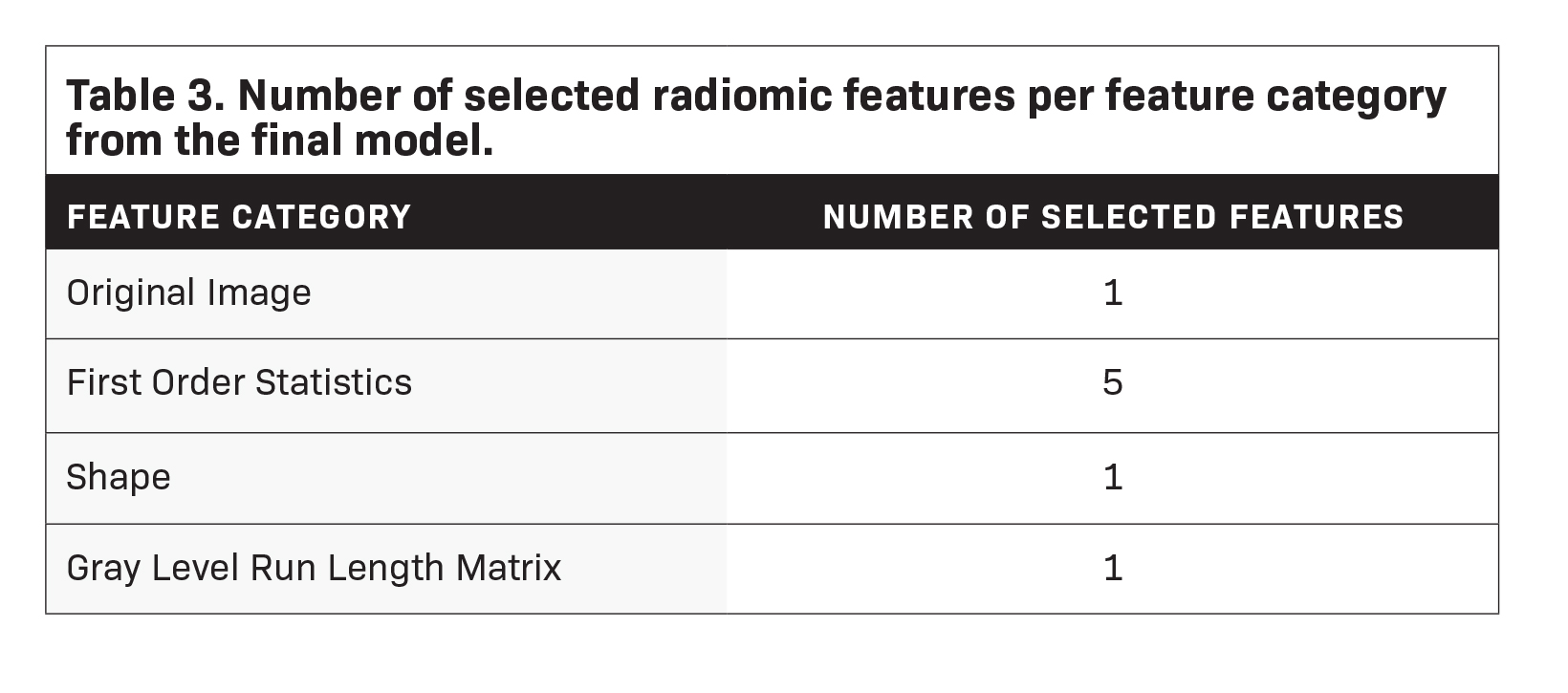
Feature importance metrics are delineated in Table 2, and corresponding feature rankings are visually represented in Figure 3; eight features were found to significantly contribute to the model. Subcategories of features and their respective counts in the final model are outlined in Table 3. The majority of the chosen features were first-order statistics (N=5). The categories of original images values, shape, and gray level run length matrix each contributed one significant feature as shown in Table 3. The feature with the greatest permutation importance was gray level run length matrix run entropy (Permutation Importance=0.223).
Discussion
This study investigated the efficacy of a radiomics-based machine-learning model to accurately determine the chronicity of pulmonary nodules in patients with NTM-LD. The model had a significantly greater AUC for the classification of acute vs. chronic pulmo- nary nodules than any of four individual radiologists. The model also had overall high performance in the classification of chronic nodules with PPV of 0.88 and moderate performance in the classification of acute nodules with NPV of 0.75. These findings under- score the potential of radiomics in predicting the future behavior of NTM-LD based on complex imaging patterns.
The variability in predicting nodule chronicity between human readers and the AI algorithm suggests an avenue for further investigation into human interpretive patterns. Specifically, the important features extracted from the radiomics analysis may provide insight into features that humans look for in determining nodule chronicity.
Radiomic features may be categorized into various groups depending on the nature of image transformation algorithms and other techniques to identify high-dimensional patterns. The Image Biomarker Standardisation Initiative has outlined a widely recognized set of radiomic features, which were used in this study.8 The most salient features in this context were first-order features, which describe basic distributions of voxel intensities within a region of interest. In this case, first-order features such as the range of voxel intensities, minimum and maximum voxel values, etc. constituted the majority of important features to the model. Shape and original image values were also of importance. Human expert readers are able to discern shape-based, original, and first-order features.9 Only one of the eight significant features was a high-dimensional feature (gray level run length matrix run entropy), being a feature not generally perceptible to humans. However, this high-dimensional feature was the single most important feature to the model as determined by permutation importance. Taken together, the fact that the majority of significant features to the model were perceptible by humans but that the most important feature was high-dimensional, indicates that there may be trends in the data imperceptible to humans, which may have implications on disease prognostication and possibly treatment.
Prior studies have employed deep convolutional neural networks (DCNNs) for NTM-LD prognostication,10 but these are often limited by their “black-box” nature. Radiomics-based models, in contrast, provide a degree of interpretability by leveraging explicit radiomic features. For example, DCNNs have successfully been applied to predict outcomes such as mortality and differentiating between nontuberculous mycobacteria and Mycobacterium tuberculosis.11 Similarly, radiomic-based analyses have been successfully applied to the prediction of NTM versus M. tuberculosis, with AUCs greater than 0.84, but have been able to identify specific features (both human interpretable and non-interpretable) to explain these predictions.12 These results from the literature highlight the ongoing clinical applications of machine learning in the management of NTM-LD and related disorders and reiterate the point that high-dimensional features predictive of relevant pathologies exist which may be imperceptible to humans.
Our study has several limitations worth noting. First, this study, as with most radiomics-based approaches, required semi-automated image segmentation for data acquisition. Semi-automated segmentation has inherent subjectivity, which can affect accuracy, though we partially compensated for this by having two readers segment the same nodules and extracted the overlapping regions for radiomics analysis. Additionally, segmentation is a labor-intensive process, which can lengthen the time required to create a useable data set in comparison to DCNN techniques, wherein a single user can process orders of magnitude more data in the same amount of time, assuming these data are available. This data set primarily includes patients who sought care at a single tertiary care center in the United States. This may limit generalizability and, more specifically, may reduce the stability of the significant features to the model. A different patient population or scan technique may identify different significant features than the ones we identified. The representation of each nodule in our dataset is limited to a single axial CT slice rather than a 3D dataset, restricting our ability to fully capture the nodule’s morphology and texture.
Our analysis is further limited to patients with follow-up CT scans. Patients with follow-up scans are generally either those with unresolved/worsening symptoms and/or those with the means to follow up with their exams. Lastly, we did not have pathologic confirmation for all of the detected nodules, adhering to the standard of care for NTM-LD surveillance, which generally reserves biopsy or surgery for specific refractory lesions or suspected malignancies.
Multiple future directions from this research may be considered. In a broad scope, one may envision a real-time probability indicator tool integrated into PACS systems for the radiologist and clinician’s use. A nodule could be segmented and the future behavior of the nodule predicted with probability scores provided for clinical reference. To reach such a point, a larger, ideally multi-institutional, dataset would be required, and the final layers of the ensemble model could be retrained on data native to specific institutions to enhance per-institution model reliability. Additionally, integration of clinical and demographic information into the model may be of use. An approach such as this, integrating radiologic and clinical data in a systematic fashion, could help improve clinical decision making in NTM-LD.
In conclusion, our radiomics-based model shows promising utility for differentiating between acute and chronic pulmonary nodules in NTM-LD patients. Its diagnostic performance was comparable to that of experienced radiologists, suggesting its value as a diagnostic adjunct. Such tools could ultimately improve clinical decision-making and patient outcomes in the management of NTM-LD.
References
- Haworth CS, Banks J, Capstick T, et al. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). Thorax. 2017;72(Suppl 2):ii1-ii64.
- Lam PK, Griffith DE, Aksamit TR, et al. Factors related to response to intermittent treatment of Mycobacterium avium complex lung disease. Am J Rresp Crit Care Med. 2006;173(11):1283-1289.
- Wang Y, Lin A, Lai Y, Chao T, Liu J, Ko S. The high value of high-resolution computed tomography in predicting the activity of pulmonary tuberculosis. Int J Tuberc and Lung Dis. 2003;7(6):563-568.
- Wang Y, Shang X, Wang L, et al. Clinical characteristics and chest computed tomography findings related to the infectivity of pulmonary tuberculosis. BMC Infect Dis. 2021;21(1):1-7.
- Liu C-J, Tsai CC, Kuo L-C, et al. A deep learning model using chest X-ray for identifying TB and NTM-LD patients: a cross-sectional study. Insights into Imaging. 2023;14(1):1-12.
- Van Griethuysen JJ, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104-e107.
- Erickson N, Mueller J, Shirkov A, et al. Autogluon-tabular: Robust and accurate automl for structured data. arXiv preprint arXiv:200306505. 2020;
- Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative-feature definitions. arXiv preprint arXiv:161207003. 2016;10
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563-577.
- Andrew C. Lancaster B, Mitchell E. Cardin B, Jan A. Nguyen M, et al. Utilizing deep learning and computed tomography to determine pulmonary nodule activity in nontuberculous mycobacterial lung disease patients. J Thorac Imag. 2023;(Article in Press)
- Xing Z, Ding W, Zhang S, et al. Machine learning-based differentiation of nontuberculous mycobacteria lung disease and pulmonary tuberculosis using CT images. BioMed Res Internat. 2020;2020
- Yan Q, Wang W, Zhao W, et al. Differentiating nontuberculous mycobacterium pulmonary disease from pulmonary tuberculosis through the analysis of the cavity features in CT images using radiomics. BMC Pulm Med. 2022;22:1-12.
Citation
TI M, C H, A L, M U, D O, H B, CT L. Automated Machine Learning with Radiomics for Predicting Chronicity of Pulmonary Nodules in Patients with Nontuberculous Mycobacterial Lung Infection. Supplement to Applied Radiology. 2024;(1):4-10.
January 25, 2024